Session March 2022: Cell economic principles in Synthetic Biology
Talk “Cell economics: the interplay between growth and gene expression in bacteria” by Jose Jimenez.
Today we were joined by Jose Jimenez, senior lecturer in Synthetic Biology at the Department of Life Sciences at Imperial College London. The group of Dr. Jimenez focuses on the connection between synthetic biology and evolution, especially as applied to understanding the tradeoffs involved in gene expression. They have more recently been using principles of economics to investigate this interplay, and today Jose Jimenez shared his group’s application of these strategies to bacterial model systems.
The talk began with the description of a paradigm familiar to any synthetic biologist - the classic “design-build-test” cycle. A typical implementation of this cycle could involve first designing a genetic circuit, assembling it and integrating it into a living system (e.g. a bacterium), and finally iterating on its design by testing its performance. This structure forms the basis for integrating new functions into living systems and reflects the roots of synthetic biology in engineering disciplines.
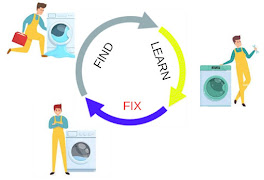
However, this paradigm is far from realistic applications. Instead, he has opted for what he termed the “washing machine repair cycle” - a structure centered on developing tools to fix a perceived problem. Here, a problem is first identified, then systems are developed to find out how the broken machine works, and then tools are used to fix it. For Dr. Jimenez, this represents a better way to approach questions of economic tradeoffs in cells, as it allows his group to more precisely center their efforts on the limitations of their internal machinery. As such, he structured his talk on the three general steps of this paradigm: Find, Learn, and Fix.
Find: What are the limitations in the gene expression machinery?
Dr. Jimenez was first interested in understanding how the cell distributes its resources to make a genetic circuit function. To do this, his group designed a system that introduced a synthetic induction cascade into bacterial Experiments were designed to quantify protein cost and understand competition for limited resources in the cells. This circuit has GFP as its output, whose expression can be tuned by modifying the concentration of its inducer molecule, the strength of its ribosomal binding site (RBS), or the number of plasmids present in each cell. In computational simulations of this system, they predicted that a high amount of GFP expression could be reached with a lower amount of inducer molecule. When they tested this system experimentally, however, they found instead that the circuit would fail if the concentration of the inducer molecule was too high. “This is clearly a problem of economics,” figured Dr. Jimenez, inferring that the cost of making a protein itself could affect circuit performance.
To look into these machinery limitations further, the group designed a simple set of experiments to quantify the cost of the proteins that make up the induction cascade. These required slightly modifying the genetic circuit in question: now, the GFP would be expressed constitutively, but an RFP was introduced and placed under the control of an inducer molecule. When the bacteria containing these circuits were cultured in vitro, the group found that GFP expression decreased as RFP increased. This in itself was expected, but what surprised them was the relationship was linear, a trend that led them to more precisely frame their observations with economic theory. Specifically, this trend resembled an isocost line, which in economics is a relationship that describes combinations of resources that produce the same output. This framing allowed Dr. Jimenez to better understand the cost tradeoffs going on in the cells: like in economics, if the slope of these isocost lines are shallower, it means that the proteins in questions are cheaper to produce. Conversely, a steeper line would indicate a more costly set of machinery.
Learn: What is the precise role of these limitations in success/failure of exogenous circuits/pathways?
In order to further understand how the translational capacity of the cell was being allocated to the function of the signaling cascade, Dr. Jimenez and his group designed a chemostat experiment in collaboration with scientists at the University of Warwick. They focused on the role of the ribosome as “the machine that makes more machines'' - a framing that centered the cell’s translational machinery squarely within his “find-learn-fix” paradigm. This set of experiments, which involved modulating the carbon sources available to the bacteria (and therefore their growth rates), led to further observations that shed light on how the cell allocates resources under differing growth conditions. For one, they observed that when the growth rate increased, few resources were allocated into making the synthetic circuit’s proteins. They looked further into this tradeoff by removing rRNA operons to decrease the cell’s ribosome budget. Under these more resource-limited conditions, they found that the identity of the available carbon source affected the budget, but not the cost or total amount of ribosomes.
These observations suggested a more general theory of "cell capacity" and growth, whereby slower growth reduces the maintenance cost of the cell and increases the budget for other proteins (e.g. from a synthetic circuit). Conversely, if the amount of rRNA available is reduced, the capacity of the system is also reduced. Using a poor carbon source in this context can then be thought of not as changing the capacity of this ‘engine,’ but rather causing it to run with a fuel source that is less efficient.
Fix: Developing strategies to improve circuits and strains such that the competition for this machinery decreases
Engineering ribosomal partitions with an orthogonal 16S RNA, it is demonstrated that ribosomal partitions alleviate couplings in pathways. Also, bet-hedging increases population fitness under stress.
Conclusions
- Isocost lines provide a conceptual framework to study microbial cell physiology.
- Chemostats can help dissect the interplay between transcription and translation.
- Pools of orthogonal ribosomes can mitigate couplings in gene expression.
- Non-essential functions can be targeted to optimize protein/pathway yields in a directed and a random way.
Discussion
Interesting perspectives were raised by the panel:
- Economic principles should be taking into account in standardizing synthetic biology
- Yeast (not same limitations are observed)
- Role of models
- Leading to further discussion on the following open questions:
- How universal are the principles that we can infer in E coli? What can we assume as general principles? How to infer them?, cell objectives, constraints
- What are the more convenient modeling frameworks to do so? Are the current modeling frameworks appropriate? Do we need “more simplified” models of metabolism? How can we get reduced models of metabolism? Do we need to take into account the dynamics of metabolism?
References
- Qian, Yili, et al. “Resource Competition Shapes the Response of Genetic Circuits.” ACS Synthetic Biology, vol. 6, no. 7, 3 Apr. 2017, pp. 1263–1272, 10.1021/acssynbio.6b00361. Accessed 8 Mar. 2022.
- Gyorgy A, Jiménez JI, Yazbek J, Huang HH, Chung H, Weiss R, Del Vecchio D. Isocost Lines Describe the Cellular Economy of Genetic Circuits. Biophys J. 2015 Aug 4;109(3):639-46. doi: 10.1016/j.bpj.2015.06.034. PMID: 26244745; PMCID: PMC4572570.
- Darlington APS, Kim J, Jiménez JI, Bates DG. Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes. Nat Commun. 2018;9(1):695. Published 2018 Feb 15. doi:10.1038/s41467-018-02898-6
Thanks to
- Speaker: Jose Jimenez (Imperial College)
- Panel: Jakob Ruess (INRIA, Pasteur Institute), Jesús Picó (UPV), Elad Noor (Weizmann Institute)
- Organizer: Irene Otero-Muras (i2sysbio, UV-CSIC)
- Bloggers: Alan Pacheco, Christian Fernández, Hamza Faquir, and Irene Otero-Muras

Comments
Post a Comment